Learning how to build a serverless deployment pipeline on Google Cloud Platform
By Lynn Langit.Apr 26, 2019
Several years ago I worked with a team to build a production CI/CD pipeline for an AWS project. At that time, none of the major cloud vendors offered their own CI/CD services, so we then used the open source GO CD server.
Although we were successful, the project was tedious and took several months. For this reason, I’ve been interested to try out integrated alternatives as they become available.
Lately, much of my production work has involved constructing and testing cloud-based data pipelines at scale for bioinformatics research. I have been thinking for awhile now about how best to use new cloud-based CI/CD services in automating these complex pipeline deployments.
Picturing Automated Deployment
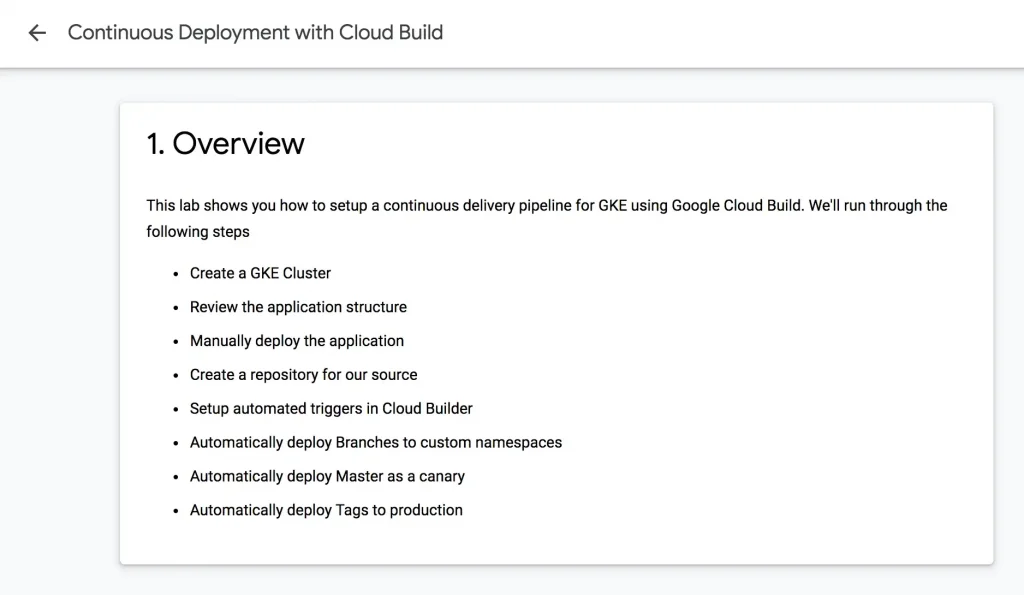
As I worked through a well-written tutorial on GCP CI/CD today, I was surprised to be unable to find any sort of diagram of the process. Although I was able to complete the tutorial successfully, I was still left feeling like I was not fully comprehending the solution architecture applications.
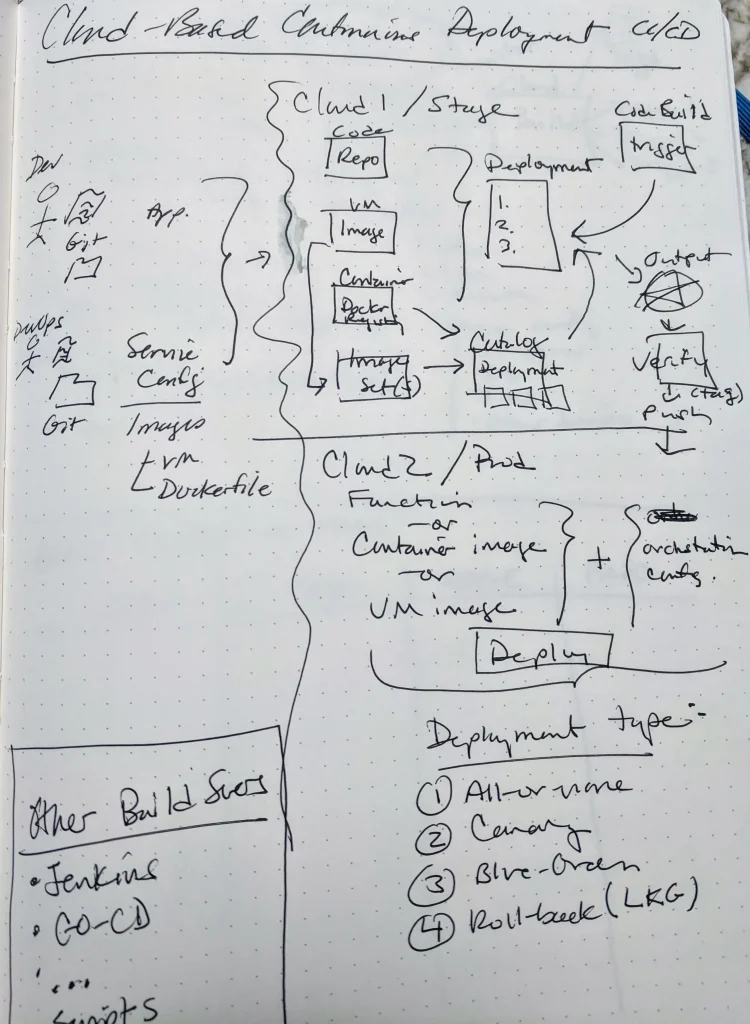
Next, I attempted to draw my own version of what I completed in the tutorial. Quickly sketching considerations from my production experience and trying to combine that with what I had learned from completing the tutorial was unsatisfying. The result is shown below.
A clear path through the complexity of the steps involved wasn’t yet apparent to me. After sketching, I tried thinking about each service or set of services that would be involved in building this type of pipeline.
Considering GCP Services
I started by trying to understand the GCP Tool Services offerings and how each service would relate to building a CI/CD solution for the type of data pipelines my teams have been building.
There are a number of tools. Below is my summarized understanding of each of the GCP services listed on the Tools menu. Note that all of these services are serverless.
- Cloud Build — to create build pipelines with steps & triggers; outputs artifacts (images…)
- Cloud Scheduler — to schedule cron jobs
- Cloud Tasks — to schedule explicit async tasks
- Container Registry — to host public or private container (docker) images
- Source Repositories — to host or sync Git code repositories
- Deployment Manager — to deploy infrastructure from static or dynamic templates
- Private Catalog — to specify approved deployments
Learning More
Then I watched a few presentations from the recent GCP:Next conference. The most useful to me were the following:
- “CI CD Across Multiple Environments” — interesting, but showed combining non GCP tools (Jenkins, HashiCorp Vault…) with GCP tools, so added even more complexity to potential scenarios.
- “Develop Faster on K8 with Cloud Build” — compelling demo of Skaffold library for K8, but no end-to-end scenario was shown.
- “Develop, Deploy & Debug with GCP Tools” — most useful was demoes showing aspects of working with various services, such as the very intelligent search in Source Repositories.
Although these presentations and doing the tutorial were helpful in my understanding of how I might use these services for my customers, I still couldn’t visualize what I would actually build. To that end, I made an attempt to diagram one possible CI/CD architecture for a use case that I’ve been working on for awhile now.
Visualizing CI/CD for GCP Data Pipelines
Here’s my attempt to design a solution for one of my customers. I’ll describe the application architecture first. The use case is bioinformatics research via a specialized machine learning library for genomics, VariantSpark.
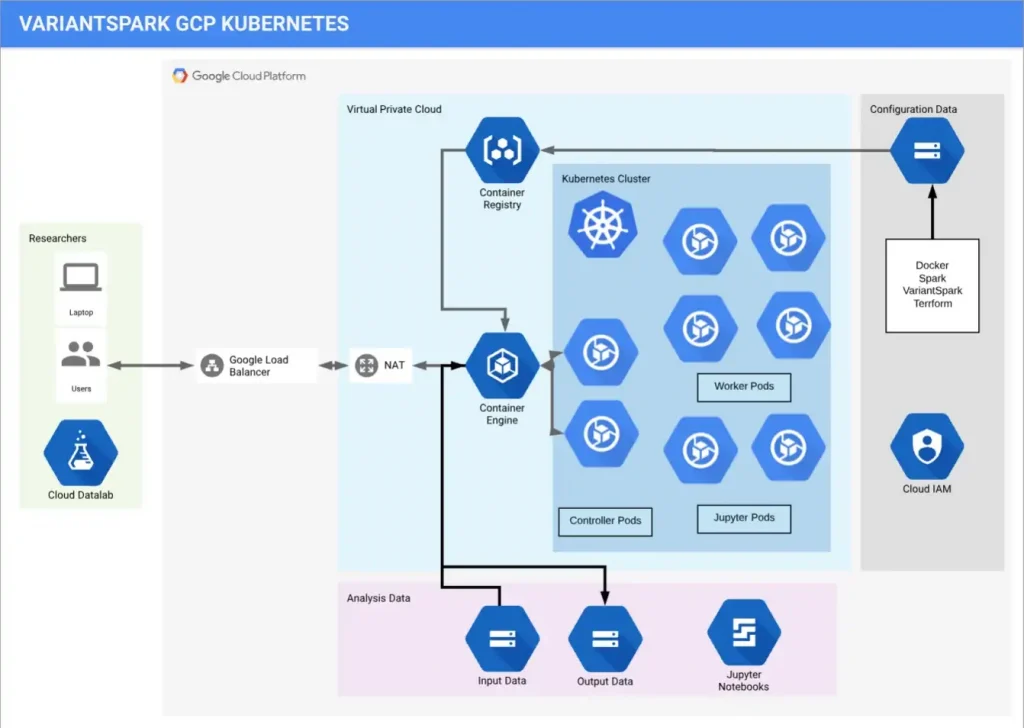
Below is a diagram of solution architecture on GCP K8:
You’ll note that this is a Data Lake with the K8 cluster processing data from GCS. Managed Jupyter Notebooks are part of the solutions as this is the preferred environment for research work.
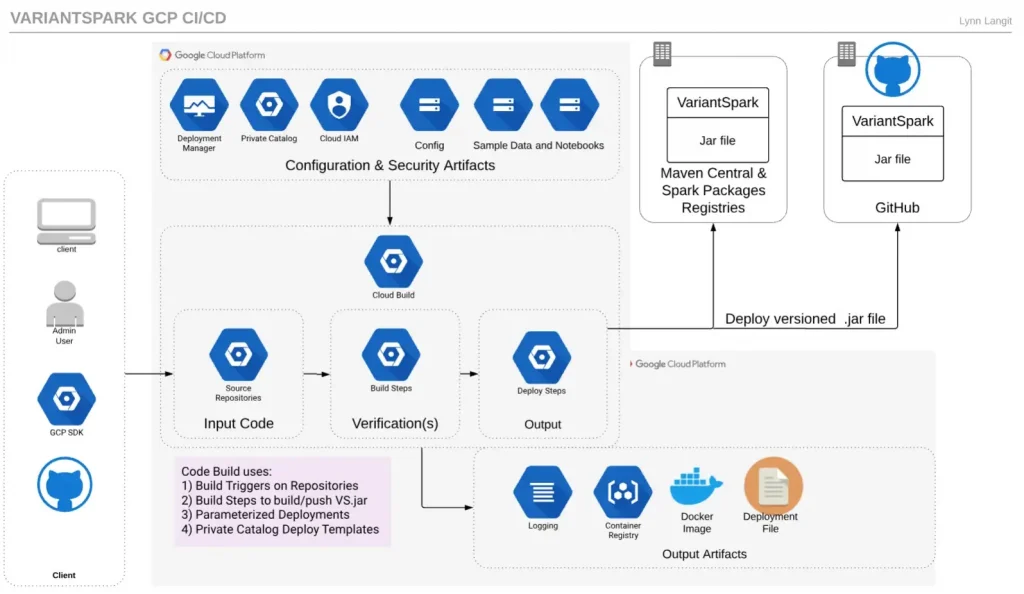
Shown below is my first draft of how I imagine my team could use GCP CI/CD services to build a pipeline to deploy a VariantSpark K8 cluster on GCP.
Because VariantSpark in an open source library, there are continual improvements to it. However, because VariantSpark is a research tool, it uses major and minor version numbers in release so that it can be cited in reproducible bioinformatics research papers.
So the process to build a cluster first requires determining which version of the VariantSpark JAR file is desired for the base container image. The pipeline may needs to perform a maven build on the source GitHub Repo for VariantSpark.
l Cloud Build artifacts include Docker Images, Kubernetes Clusters and more
GCP Source Repositories can mirror GitHub Repos. Cloud Build can use triggers on Source Repositories to trigger builds. Cloud Build includes builder (task) for various types of images including maven. Cloud Build builders also include docker and kubectl, so the VariantSpark JAR file could be built as part of the docker image and that image could be used to spawn a GKE cluster.
Additionally, GCP Deployment Manger could serve a YAML-deployment which details other GCP service configurations that are needed for a GKE cluster. It also seems possible to use GCP Private Catalog for research groups who have GCP Organizational Accounts to offer a VariantSpark GKE deployment template.
Next Steps
Now that I’ve learned the fundamentals of these services, I am looking forward to working with research and DevOps teams world-wide to build serverless CI/CD pipelines for bioinformatics.
The original article published on Medium.
Lynn Langit is a big data and cloud architect, a Google Cloud Developer Expert, an AWS Community Hero, and a Microsoft MVP for Data Platform.